Rewriting Life
Cells as Circuits
Techniques borrowed from computer engineering could help identify promising drug targets.
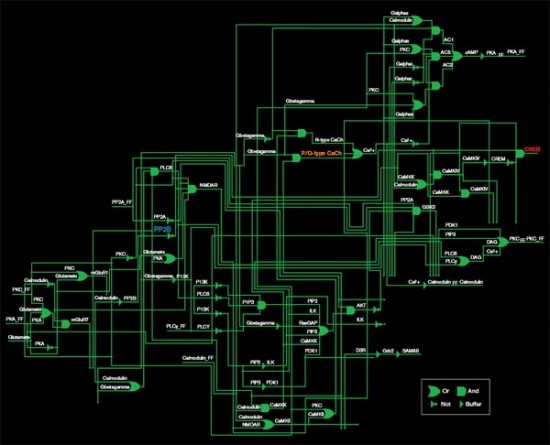

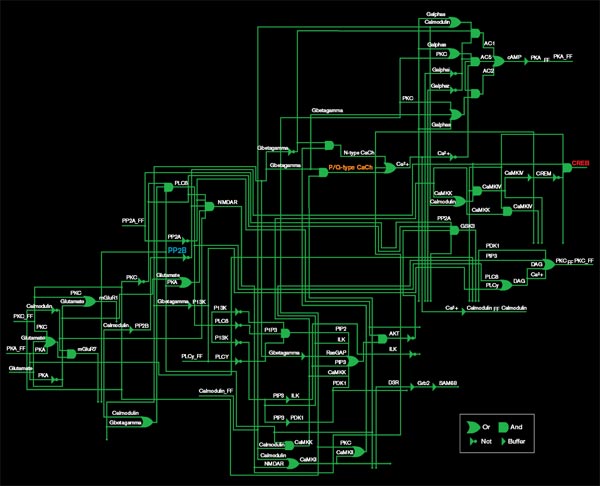
Source: Effat Emamian and Ali Abdi
The map of the human genome was expected to spark a revolution in drug discovery. But cells’ behavior is determined by the interactions of hundreds of different proteins in what are known as signaling pathways, and figuring out which interactions go awry in disease has proved much harder than people thought.
Now, a study led by scientists affiliated with the New Jersey Institute of Technology (NJIT) has used circuit analysis to identify the molecules that play the biggest roles in cells’ signaling pathways. The researchers believe that their work can save drug researchers time and money by identifying promising drug targets.
The diagram above represents only half of a pathway that regulates the production of a molecule called CREB (red), which is involved in memory formation. The pathway is represented as a set of logic gates, the basic elements of digital circuits. An OR gate, for example, produces an output if it receives either of its inputs: here, that means that either of two molecules regulates the production of the next molecule in the pathway. Though each step in the pathway has been well studied in biology labs, the diagram provides a quantitative purchase on the network as a whole.
One molecule in this network, PP2B (blue), is being investigated as a target for schizophrenia drugs. But the researchers’ analysis shows that PP2B is less important to CREB regulation than P/Q-type calcium channels (orange). PP2B plays a role in many other pathways, so targeting it could have unfortunate side effects. Calcium channels are the target of drugs already approved for other disorders, so they may also be a safer target for schizophrenia drugs.
The researchers applied their approach to three different signaling pathways, and their predictions agreed with experimental results. Effat Emamian, who led the research, has founded a company that markets signaling-pathway analyses to drug companies.